
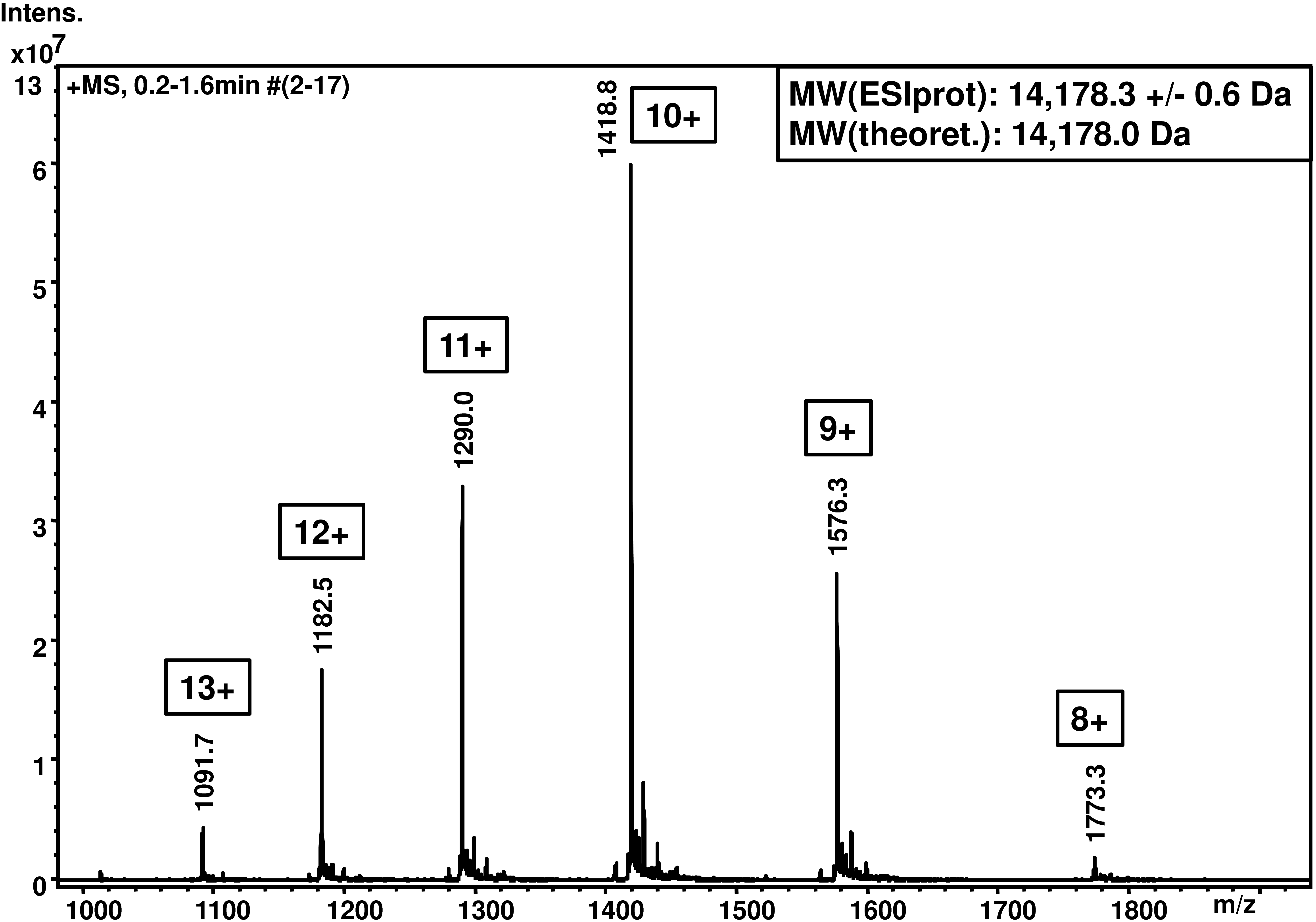
However, they do associate in discrete ratios of ions. Ionic substances are not chemically bonded and do not exist as discrete molecules. The formula weight and the molecular weight of glucose is thus: It is calculated in practice by summing the atomic weights of the atoms making up the substance’s molecular formula. For example, carbon, hydrogen and oxygen can chemically bond to form a molecule of the sugar glucose with the chemical and molecular formula of C 6H 12O 6. Molecular weight, also called molecular mass, is the mass of a molecule of a substance, based on 12 as the atomic weight of carbon-12. If a substance exists as discrete molecules (as with atoms that are chemically bonded together) then the chemical formula is the molecular formula, and the formula weight is the molecular weight. For example, water (H 2O) has a formula weight of: Humana Press.The formula weight of a substance is the sum of the atomic weights of each atom in its chemical formula. In The proteomics protocols handbook (pp. Protein identification and analysis tools on the ExPASy server. Gasteiger, E., Hoogland, C., Gattiker, A., Wilkins, M. The formula and amino acid scale are the same available on ExPASy Compute pI/Mw tool: For average weight, the ExPASy tools use the following mass scale:, while UniMod and Mascot use a slightly different one. In Peptides the function mw computes the molecular weight using the same formulas and weights as ExPASy's "compute pI/mw" tool (Gasteiger et al., 2005). The molecular weight is directly related to the length of the amino acid sequence and is expressed in units called daltons (Da). The molecular weight is the sum of the masses of each atom constituting a molecule.
#MOLECULAR WEIGHT CALCULATION TOOL CODE#
Use the amino acid one letter code as names and the mass shift in Dalton as values. Overwrites input in aaShift.ĭefine the mass difference in Dalton of given amino acids as a named vector. Set the mass scale to use for average weight only (if 'monoisotopic = FALSE'). Mw ( seq, monoisotopic = FALSE, avgScale = "expasy", label = "none", aaShift = NULL )Ī logical value 'TRUE' or 'FALSE' indicating if monoisotopic weights of amino-acids should be used
#MOLECULAR WEIGHT CALCULATION TOOL SERIES#

massShift: Calculate the mass difference of modified peptides.lengthpep: Compute the amino acid length of a protein sequence.kideraFactors: Compute the Kidera factors of a protein sequence.instaIndex: Compute the instability index of a protein sequence.hydrophobicity: Compute the hydrophobicity index of a protein sequence.hmoment: Compute the hydrophobic moment of a protein sequence.fasgaiVectors: Compute the FASGAI vectors of a protein sequence.crucianiProperties: Compute the Cruciani properties of a protein sequence.crossCovariance: Compute the cross-covariance index of a protein sequence.charge: Compute the theoretical net charge of a protein sequence.boman: Compute the Boman (Potential Protein Interaction) index.blosumIndices: Compute the BLOSUM62 derived indices of a protein sequence.autoCovariance: Compute the auto-covariance index of a protein sequence.
aaList: Return a vector with the 20 standard amino acids in upper.aaDescriptors: Compute 66 descriptors for each amino acid of a protein.AAdata: Properties, scales and indices for the 20 naturally occurring.aaComp: Compute the amino acid composition of a protein sequence.


 0 kommentar(er)
0 kommentar(er)
